Modeling the interaction between proteins and ligands and accurately predicting their binding structures is a critical yet challenging task in drug discovery.
Recent advancements in deep learning have shown promise in addressing this challenge, with sampling-based and regression-based methods emerging as two prominent approaches.
However, these methods have notable limitations. Sampling-based methods often suffer from low efficiency due to the need for generating multiple candidate structures for selection.
On the other hand, regression-based methods offer fast predictions but may experience decreased accuracy.
Additionally, the variation in protein sizes often requires external modules for selecting suitable binding pockets, further impacting efficiency.
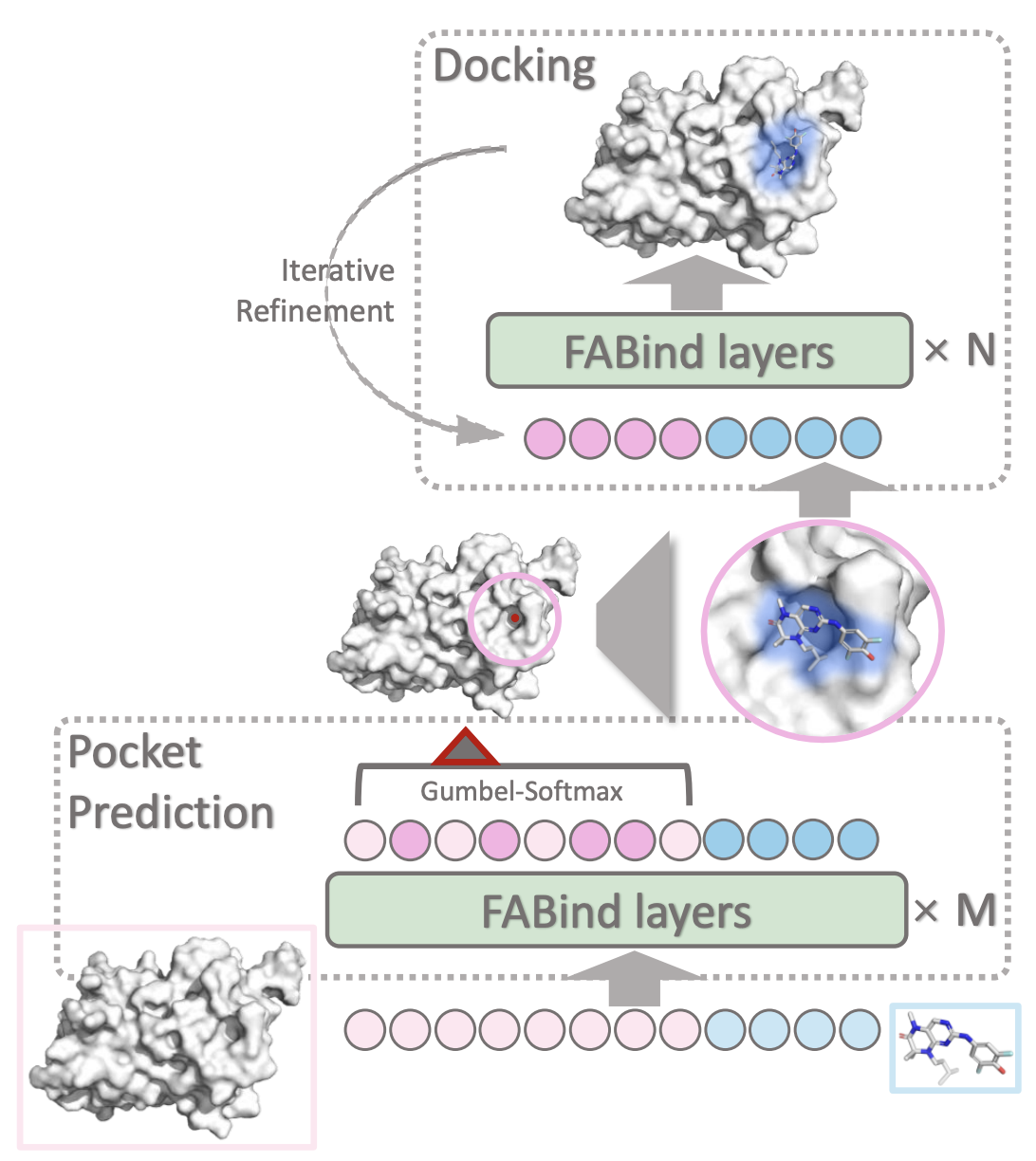
In this work, we propose FABind, an end-to-end model that combines pocket prediction and docking to achieve accurate and fast protein-ligand binding.
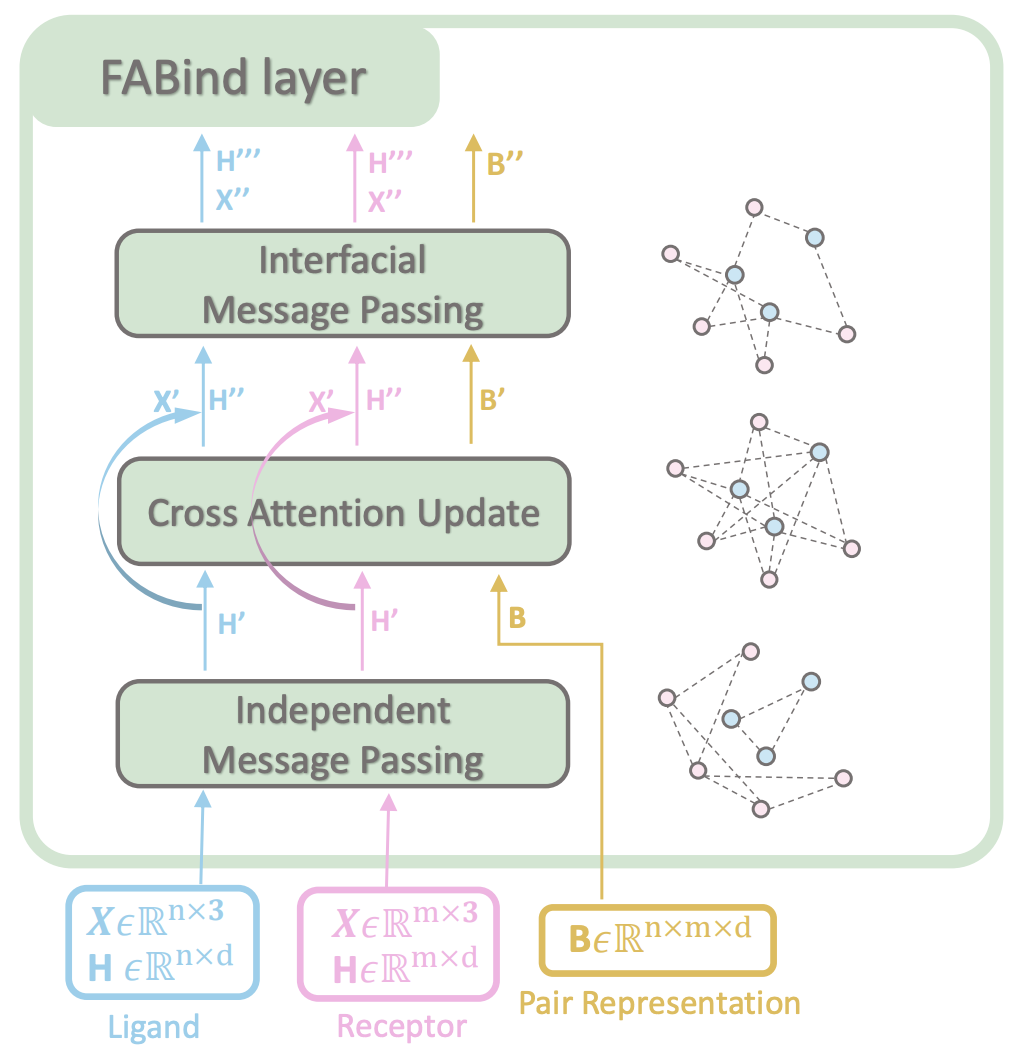
FABind incorporates a unique ligand-informed pocket prediction module, which is also leveraged for docking pose estimation.
The model further enhances the docking process by incrementally integrating the predicted pocket to optimize protein-ligand binding, reducing discrepancies between training and inference.
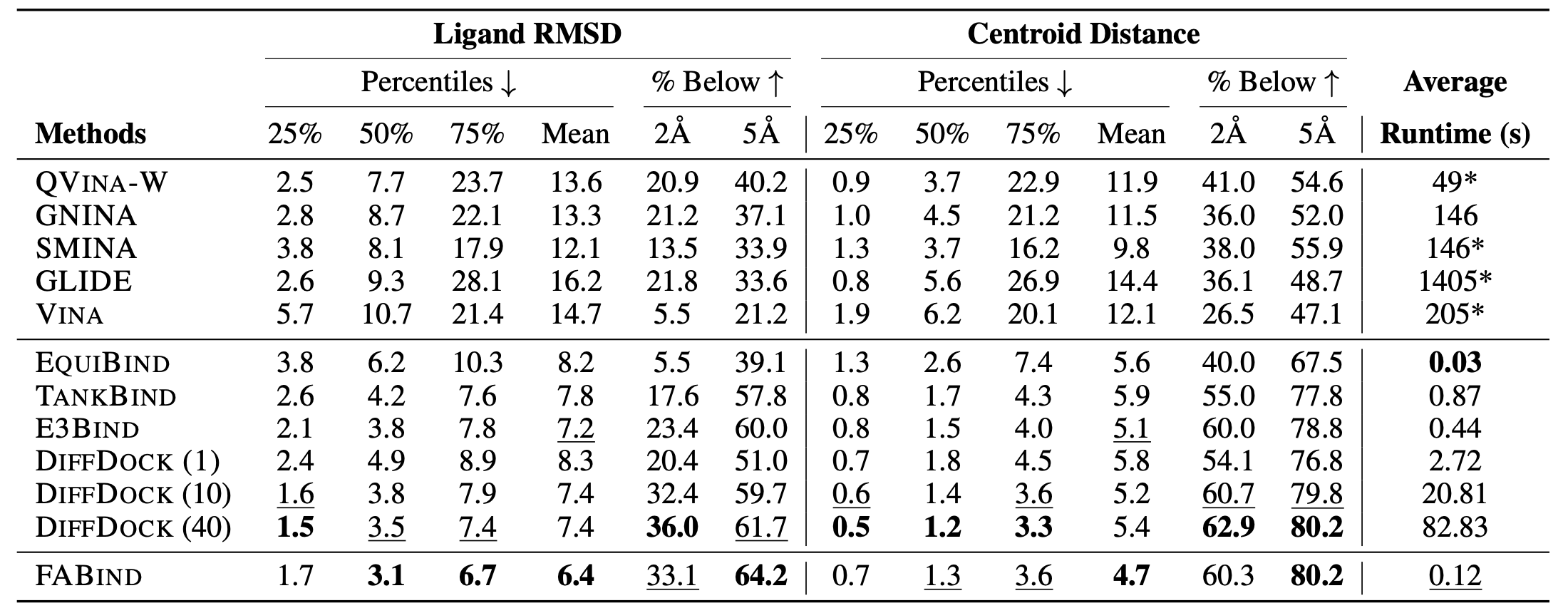
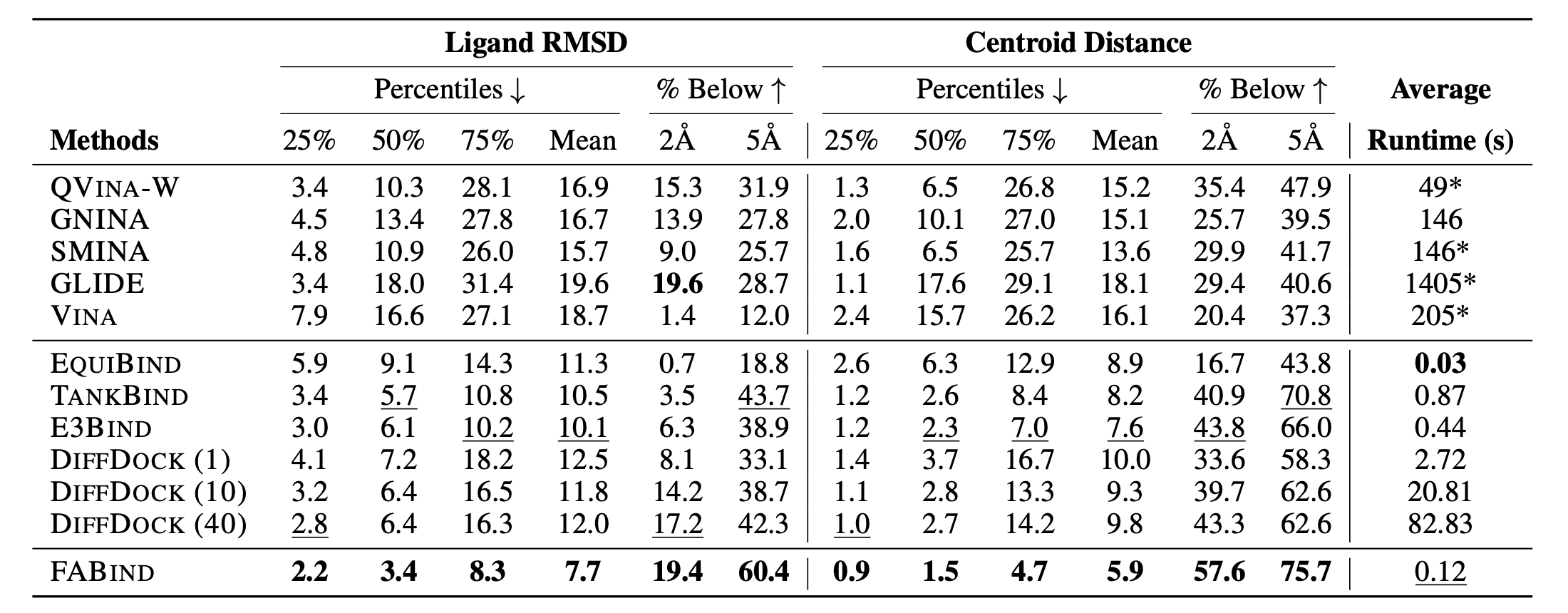
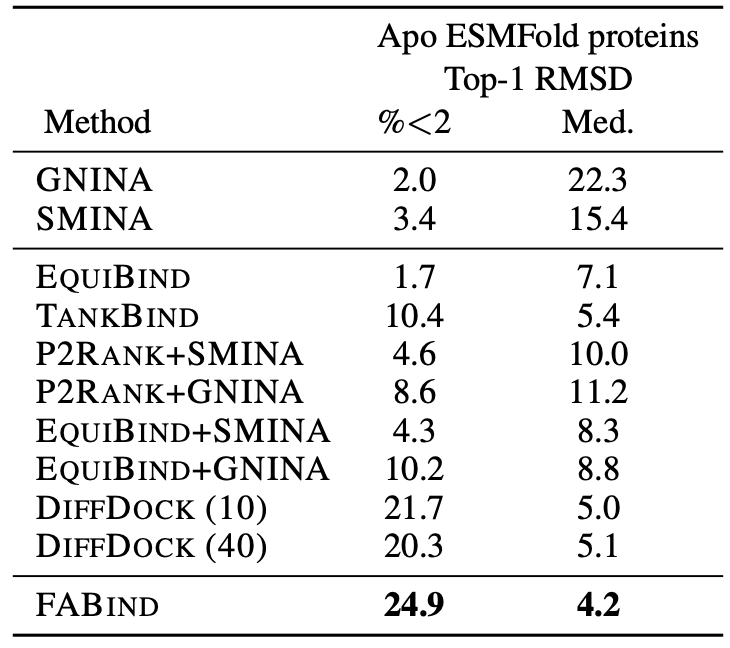
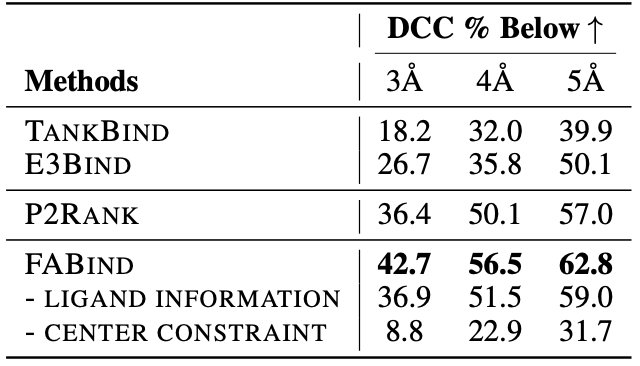
Through extensive experiments on benchmark datasets, our proposed FABind demonstrates strong advantages in terms of effectiveness and efficiency compared to existing methods.